We develop multiscale molecular models, design in silico experiments and use advanced simulation & analysis approaches to study complex molecular systems,
particularly interested in dynamic bioinspired materials, self-assembling structures and stimuli-responsive autoregulated systems.
Recent research HOT topics include:
Dynamic Supramolecular Polymers
Supramolecular polymers, where monomers self-assemble in 1D via noncovalent interations, possess unique bioinspired properties (self-healing, adaptivity, etc.). Molecular modeling can be fundamental to understand these complex materials and to promote their rational design.
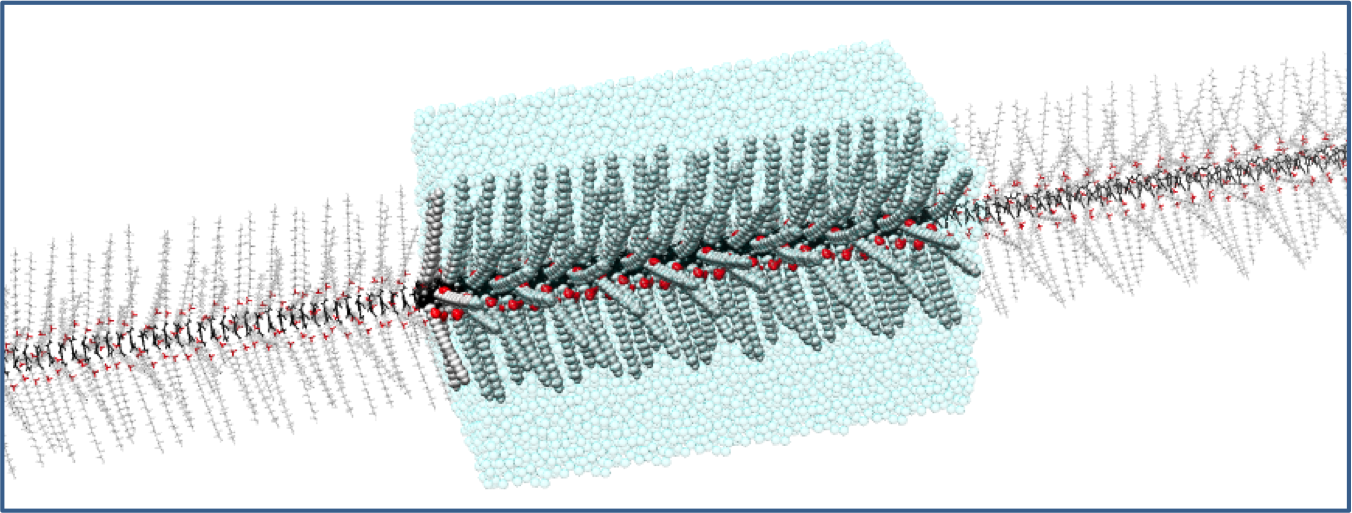
A few selected papers:
Nature Commun. 2017, 8, 147
JACS 2018, 140, 10570-10577
J. Phys. Chem. Lett. 2017, 8, 3813-9
ACS Nano 2017, 11, 1000-11
JACS 2016, 138, 13985-95
Nature Commun. 2015, 6, 6622
Adaptive and Stimuli-Responsive Materials
- Self-assembly can be used for the creation of smart and functional supramolecular materials that adapt or respond to external stimuli in controlled way. Modeling can help to understand how these work and how to design the monomers to control the adaptive properties.
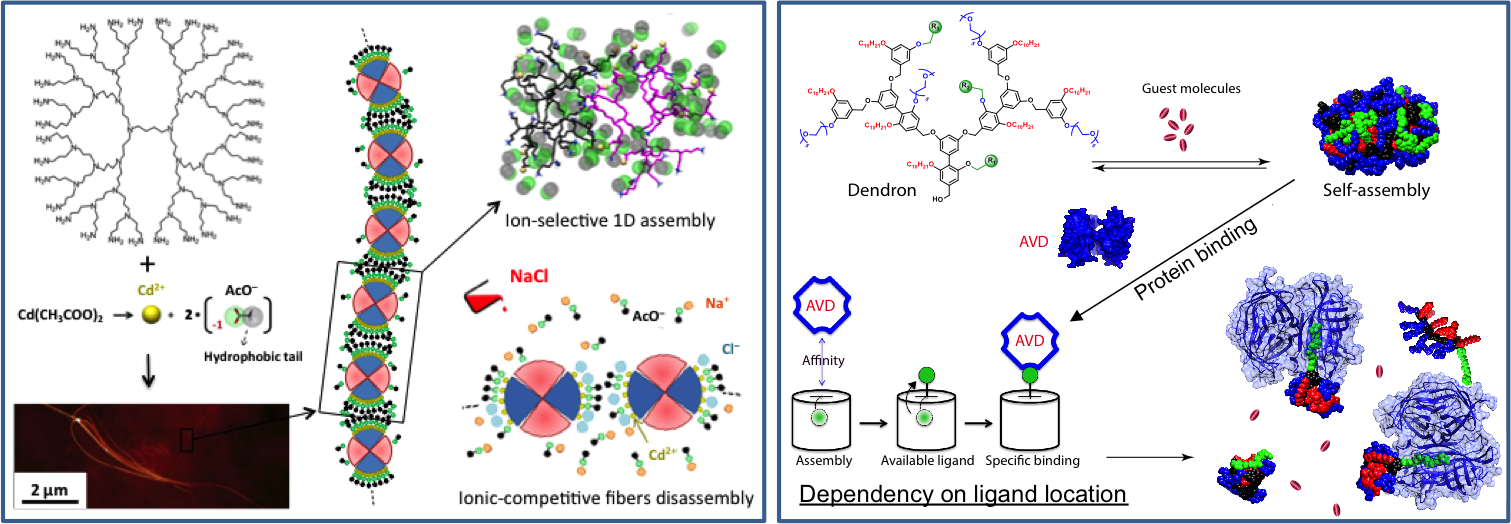
A few selected papers:
JPCB 2018, 122, 4169-4178
PNAS 2017, 114, 11850-11855
JACS 2014, 136, 5385-99
Angew. Chem. Int. Ed. 2017, 56, 4145-9
JACS 2012, 134, 3349-57
"Molecular glue" - Directed Assembly - "Molecular glue", or using molecular binders to control the assembly of other building blocks, is an intriguing way to manipulate existing nanostructures (e.g., virus, protein capsids, microtubules, etc.) and to exploit their unique properties. Molecular modeling can help the design of molecular glues to obtain new fascinating materials.
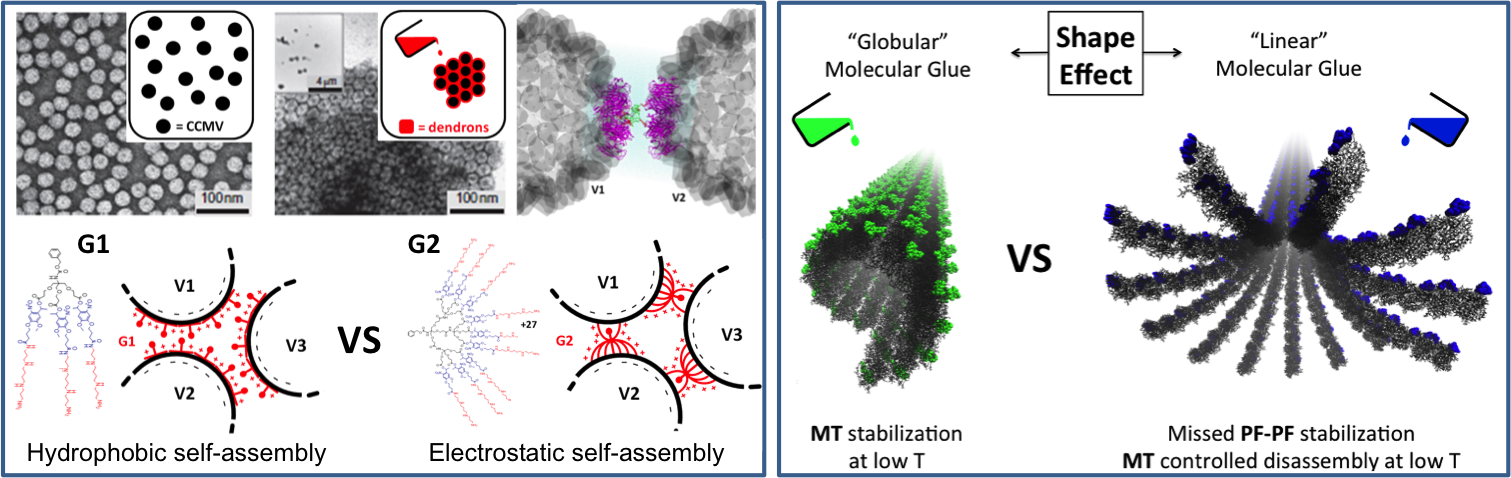
A few selected papers:
ACS Nano 2018, 12, 8029-8036
ACS Nano 2014, 8, 904-14
Nano Lett. 2011, 11, 723-8
Self-Assembled Nanostructures & Computational Nanotechnology - Soft-nanostructures and hybrid organic-inorganic systems possess interesting properties but are often difficult to characterize. Molecular simulation can be useful to explore self-assembled nanostructures (monolayers, nanoparticles, etc.), liquid crystals, nanocages, etc. where molecular crowding and interactions play a key role.
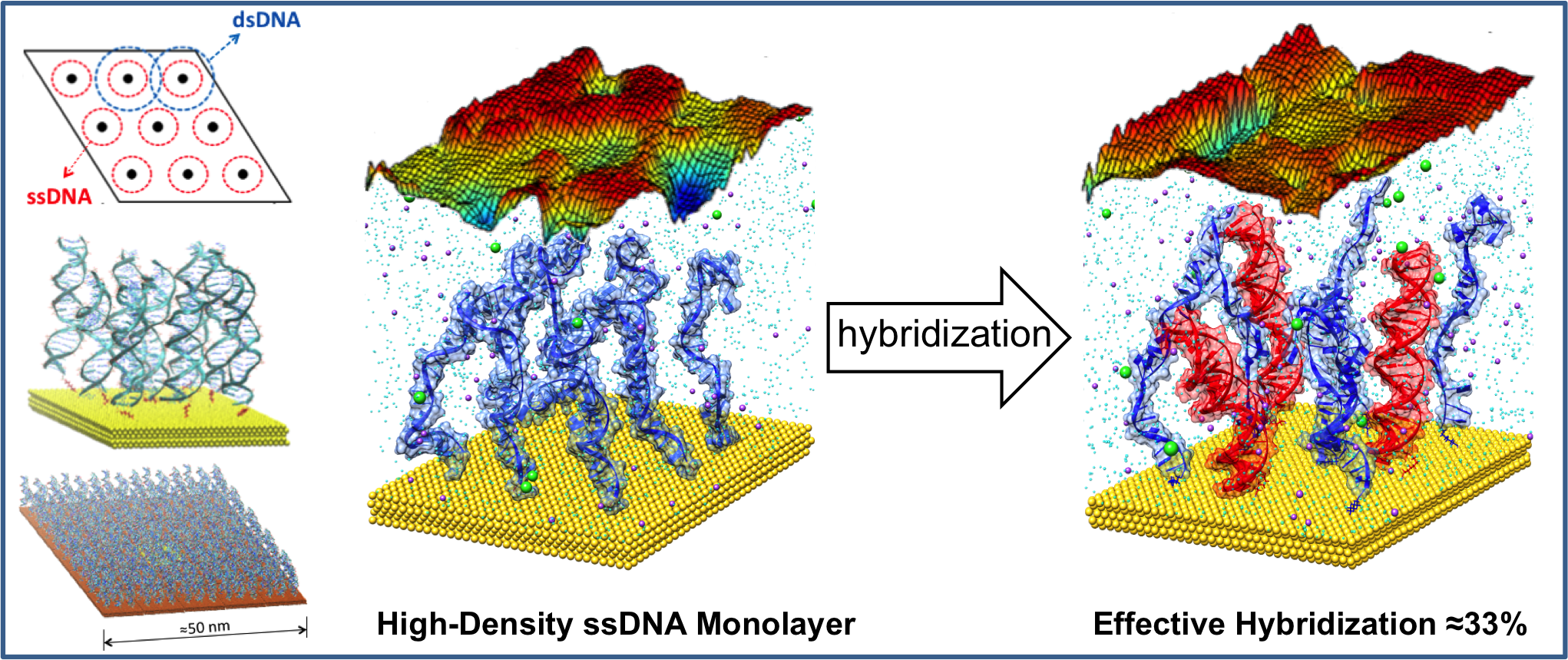
A few selected papers:
ACS Nano 2018, 12, 8029-8036
Nanoscale 2017, 9, 6399-6405
Nanoscale 2013, 5, 9988-93
Angew. Chem. Int. Ed. 2016, 55, 1685-9
Chem. Commun. 2015, 51, 1811-4
Computational Caracterization of Complex Macromolecules
The characterization of complex macromolecules in solution is a hard issue. Modeling can be a real support for the experiments providing high-resolution characterization of complex systems in solution - a "virtual microscope" providing an atomistic-level picture of the macromolecule in realistic environments.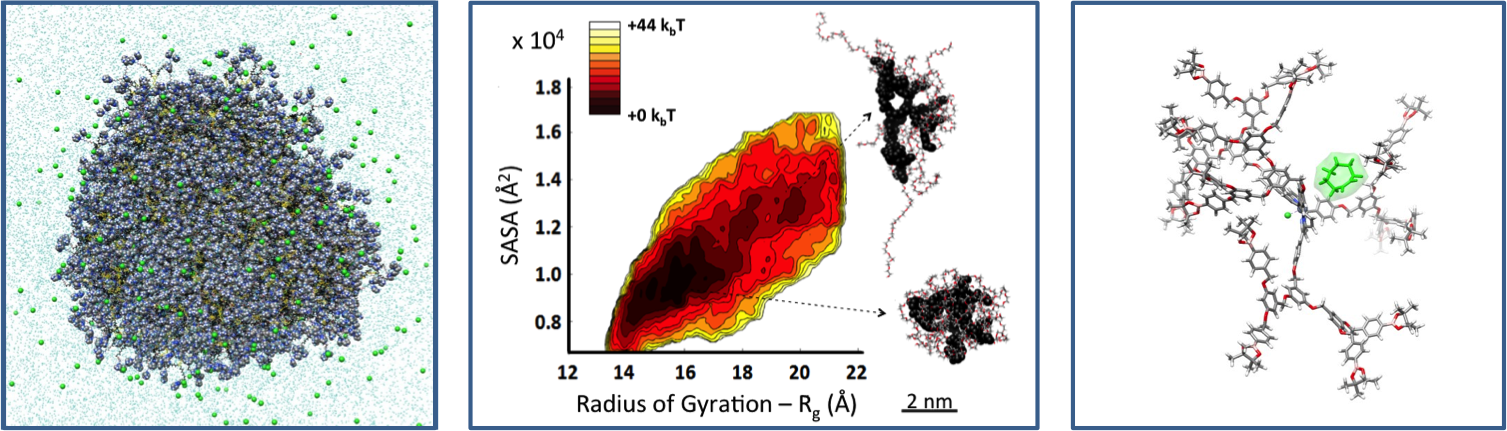
A few selected papers:
Nature Commun. 2015, 6, 7722
Soft Matter 2013, 9, 2593-7
JACS 2012, 134, 1942-5
Multivalent, Cooperative & Complex Molecular Interactions - The interaction between complex macromolecules is typically complex phenomenon, often involving binding cooperativity and multivalency. Molecular simulations can provides a privileged point of view into complex interactions, discerning the key forces/factors controlling binding.
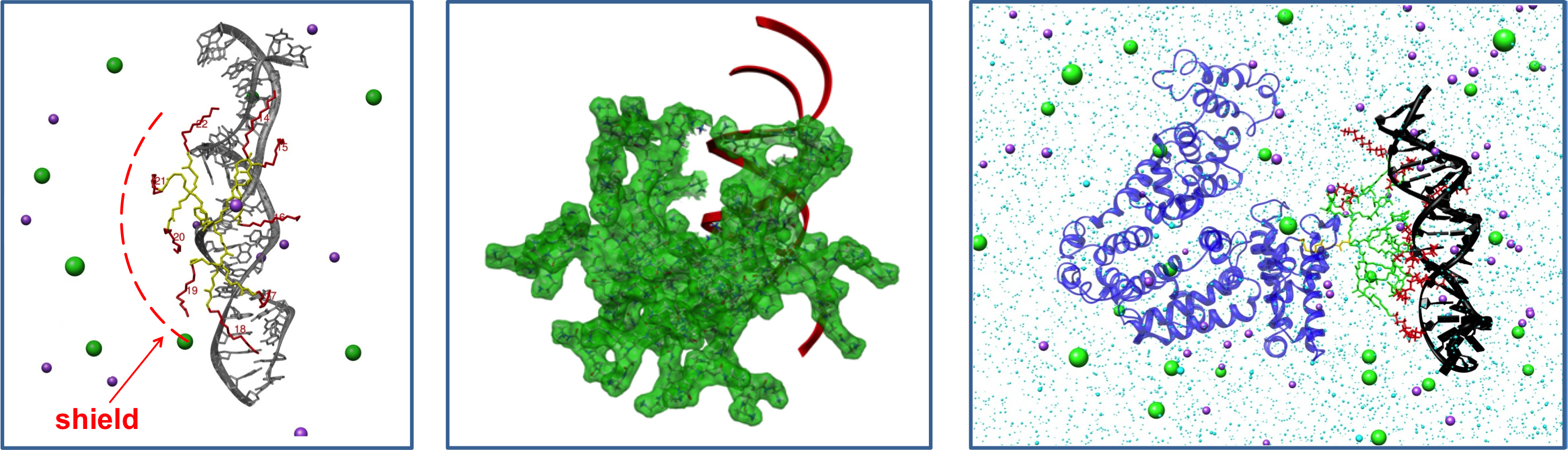
A few selected papers:
JACS 2009, 131, 9686-94
Chem. Med. Chem. 2014, 9, 2623-31
JPCB 2010, 114, 2667-75
